Conjugated Oligoelectrolyte (COE) and ssDNA for Biosening Analysis
Research Ideas:
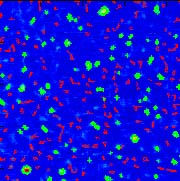
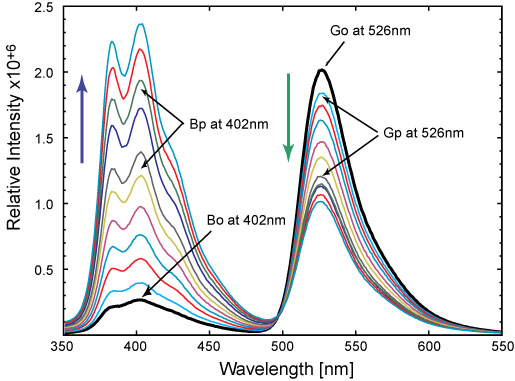
Visualization of Conjugated Polyelectrolyte (CPE) - DNA Nanoscale Aggregates
In the view of growing interest in developing independent biological assays for pathogen identification, we implemented cationic conjugated oligoelectrolyte (COE) and fluorescein labeled ssDNA molecules complex. Efficient assay for bacteria, viruses and other microorganism detection remains an important problem in health care, food safety and environmental applications. This technique can be potentially used in places where fast and reliable pathogens detection is important to prevent disease propagation, such as medical diagnosis, food and water supplies quality control. Further investigation can also lead to implementation in identification of biological warfare agent. Collaboration with Prof. Guillermo C. Bazan lab in Department of Chemistry UCSB
Conjugated polyelectrolytes (CPEs) contain a backbone with a pi-delocalized electronic structure and pendant groups bearing ionic functionalities. These structural attributes allow one to combine the semiconducting and light harvesting qualities of conjugated polymers with the behavior of polyelectrolytes, for which properties such as the degree of aggregation in water and the coil dimension/stiffness are modulated by electrostatic forces.
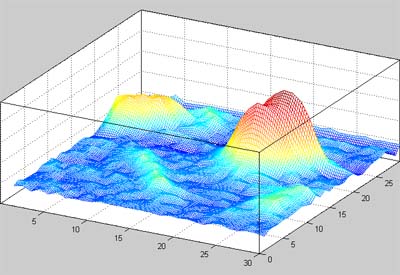
Understanding of Polymer – DNA Interactions Based on Sequential Analysis of AFM Imaging
We have developed a software tool that is able to reconstruct topological images in sequential fashion to provide precise statistical data about the composition of aggregations. Our proposed approach is based on multistage thresholding and mathematical morphology methods. Multistage thresholding allows us to segment multi-scale objects observed in AFM images. Subsequently, morphological image reconstruction provides us a way to obtain topological description of observed polymers and DNA. Collaboration with Dr. Boguslaw Obara.
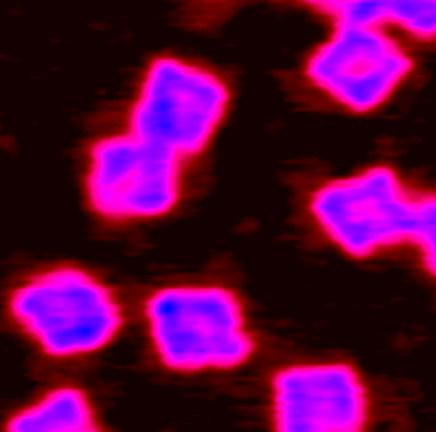
Design and Engineering of Programmable Self-assembling Materials Based on RNA and other Nucleic Acids
The incredible informative power of RNA biopolymers should allow, in theory, to direct the self-assembly of hundreds of different molecules, and to specify the precise positioning of each of these molecules within a predefined network. RNA tectonics is presently used to build programmable RNA molecules able to self-assemble into two, three and four dimensional nano-arrays. Collaboration Prof. Luc Jaeger in Department of Chemistry UCSB
Nucleic acid - Cationic Amphiphile Material
Nucleic acids are the good example of the supra-molecular materials to create complex supra-molecular structures that are well-known as a source of biological information depending on their base sequences. Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) represent the interesting biomaterials because of their rod-like structure that come from the helical structure built of electron-conductive base-pair stacking. In this study, nucleic acid / cationic amphiphile materials are prepared by exchanging the counter-ions of the phosphate moieties with cationic amphiphile. Bio-interaction between nucleic acid and cationic amphiphile is investigated to better understand these novel bioinspired materials. Collaboration with Dr. Wirasak Smitthipong from Kasetsart University, Bangkok.

Origin of Life Between Mica Sheets
Life may have begun in the protected spaces inside of layers of the mineral mica, in ancient oceans, according to a new hypothesis. The Helen Hansma mica hypothesis proposes that the narrow confined spaces between the thin layers of mica could have provided exactly the right conditions for the rise of the first biomolecules –– effectively creating cells without membranes. The separation of the layers would have also provided the isolation needed for Darwinian evolution. Dr. Helen Hansma reevaluated this idea.

1998 - 2015 AlmanaC. All rights reserved.